In order to compare how technologies handle data, I set up an ETL funnel in each environement, extracting data from Wikipedia and transforming it into a PARQUET file.
From Wikipedia
Data description: women’s profiles that are less tagged, less connected to groups, and therefore quite lost within the information system. I limited my target to the French-written part of Wikipedia
What I learned (the hard way) on Wikipedia: APIs connect to articles’ management metadata and SPARQL connect to data and categories
First step: build the SPARQL query
I won’t give a course on Wikidata’s architecture: the Wikidata Query Service offers a Query Helper to help finding the parameters for building the right query.
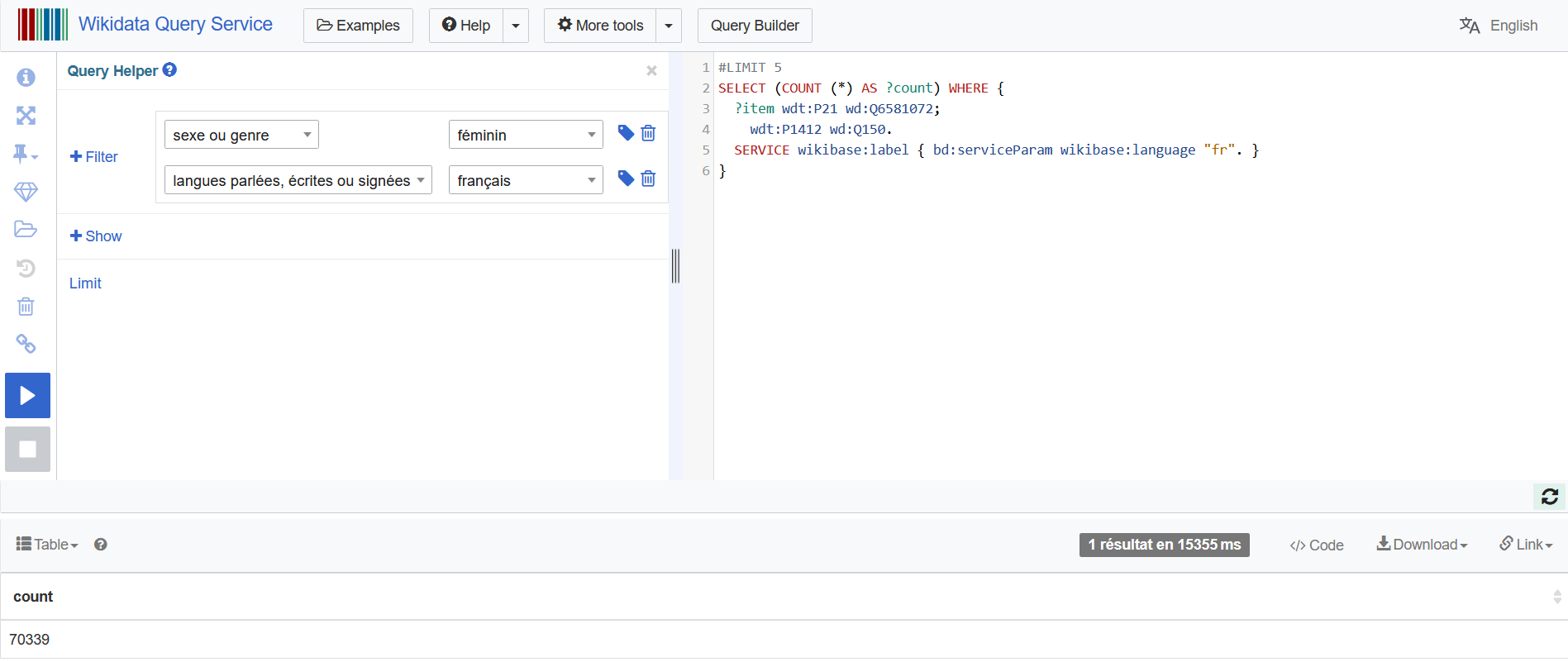
I combined :
- parameter P21 sex or gender: category Q6581072 female
- parameter P1412 languages spoken, written or signed: category Q150 French
- parameter of wikibase:language “fr”
In order to check that the query runs, it’s better to first count results.
SELECT (COUNT (?item) as ?count)
WHERE {
?item wdt:P21 wd:Q6581072;
wdt:P1412 wd:Q150.
SERVICE wikibase:label { bd:serviceParam wikibase:language "fr".}
}Result: 70349 records (view live)
Then the query can be checked with data vizualisation: let’s limit results to 100.
SELECT ?item ?itemLabel
WHERE {
?item wdt:P21 wd:Q6581072;
wdt:P1412 wd:Q150.
SERVICE wikibase:label { bd:serviceParam wikibase:language "fr".}
}
LIMIT 100Result: 100 results in 3422 ms (view live)
Second step: enrich the SPARQL query with aggregation
Let’s then aggregate the fields “has part(s)” and “part of”:
SELECT ?item ?itemLabel
(COUNT (?has_part_s_) as ?count_has_part_s_)
(COUNT (?part_of) as ?count_part_of)
WHERE {
?item wdt:P21 wd:Q6581072;
wdt:P1412 wd:Q150.
SERVICE wikibase:label { bd:serviceParam wikibase:language "fr".}
OPTIONAL { ?item wdt:P527 ?has_part_s_. }
OPTIONAL { ?item wdt:P361 ?part_of. }
}
GROUP BY ?item ?itemLabelResults: 70349 résultats en 43383 ms (view live)
NB: to get ?itemLabel appearing in the results, it has to be mentionned in GROUP BY.
Third step: generate a data feed
The Wikidata SPARQL sandbox offers many different ways of sharing:
- Sharing the query via: URL,HTML, Wikilink, PHP, JavaScript (jQuery), JavaScript (modern), Java, Perl, Python, Python (Pywikibot), Ruby, R, Matlab, listeria, mapframe
- Download results via: JSON file, JSON file (verbose), TSV file, TSV file (verbose), CSV file, HTML table
- Get a link to: Short URL to query, Short URL to result, SPARQL Endpoint, Embed result
We’ll check with each technologie what’s the best feed format to use.
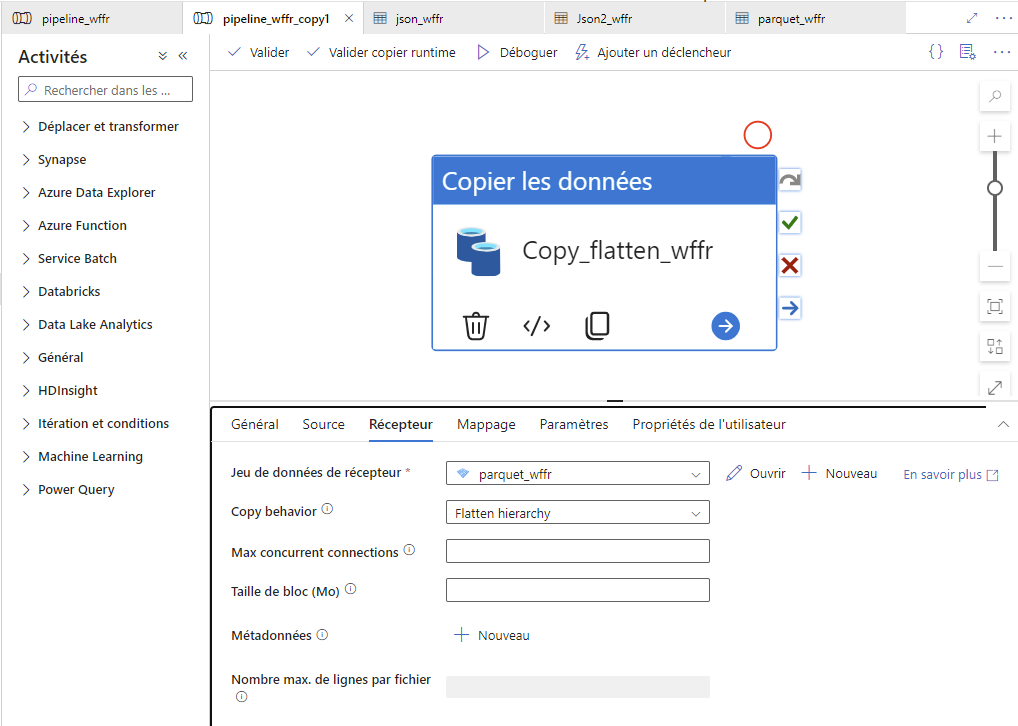
To MS Azure Data Factory
MS Azure does not handle SPARQL. It is therefore necessary to export the dataflow in another format: I chose as a JSON feed. The good practices ask to import it into a JSON file in an Azure blob storage before transforming it into another file format. Finally, I chose PARQUET as an an handier end file format.
- Create a new pipeline in Azure Data Factory
- To copy the JSON feed into a JSON file
- Mention Wikidata time limit: 60 seconds; retry after 120seconds
- Import its schema
- Source: create a new datasource with a GET connection to HTTP anonymous URL
- as base URL https://query.wikidata.org/
- as relative URL as REST in JSON format and the SPARQL query
- and a header to announce you’re not a bot; for example:
- User-Agent : AzureLearning/0.0 (https://www.aubertm.me marina@aubertm.me)
- Destination: create a storage account then a blob container to store the JSON output dataset.
- Map the source fields for the destination fields.
- /!\ Important: validate all, publish all, then import a second time the output dataset format.
Then again : validate all, publish all
Validate the pipeline, Validate runtime, Debug - Create a second pipeline in Azure Data Factory
- To copy and flatten the JSON file into a PARQUET file
- Source: the JSON file you just created
- Destination: you can use the same blob container as before.
- again :
/!\ Important: validate all, publish all, then import a second time the output dataset format.
Then again : validate all, publish all
Validate the pipeline, Validate runtime, Debug
You can visualize data from the datasets.
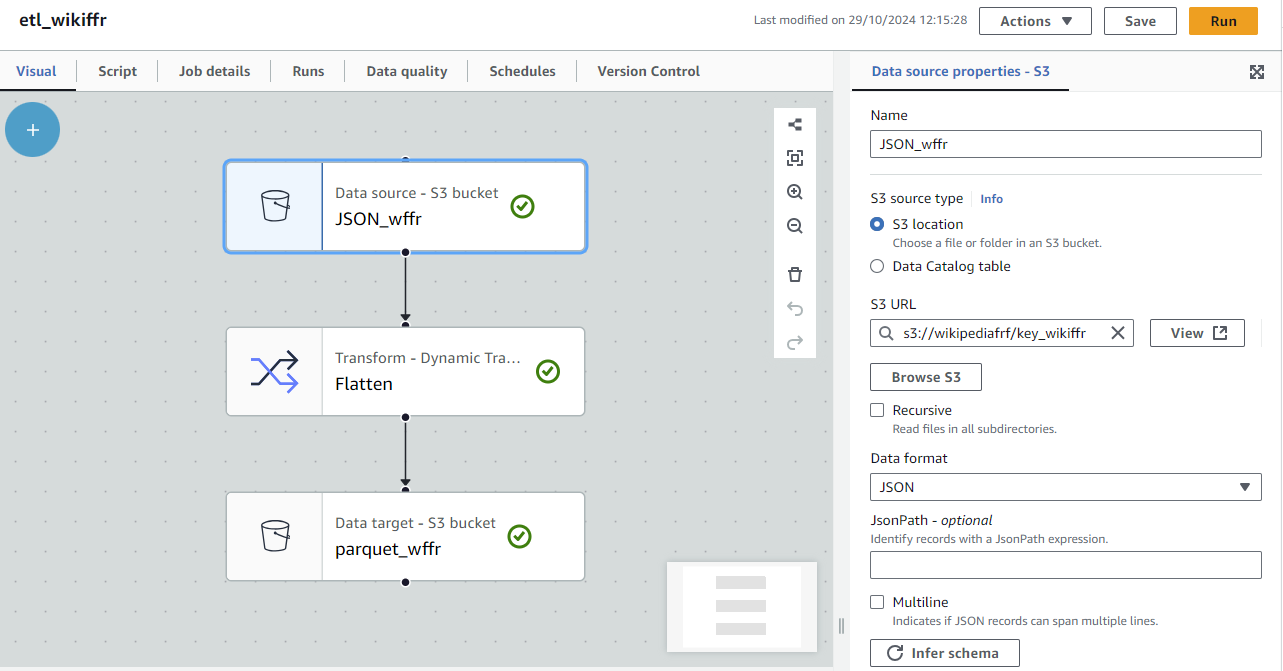
To AWS Glue
AWS does not handle SPARQL either. Let’s keep on using our Wikidata’s JSON feed.
- In AWS Glue Studio, configure IAM
- In Amazon S3 storage, create a bucket and its endpoint
- In AWS Glue Studio,
- create a Python notebook to connect the Wikidata’s JSON feed to the S3 storage space
- create a Visual ETL
- Source: select the S3 location of the JSON file you just created with the notebook; pick JSON format
/!\ leave Recursive blank - Transform: Flatten
- Target: same S3 location; pick PARQUET format
- Source: select the S3 location of the JSON file you just created with the notebook; pick JSON format
- Hit RUN
- Vizualise your new PARQUET file in S3 container
To GCP
Google Cloud Service only accepts TSV files, but then Wikidata refuses the googlebot’s request. So I had to go through BigQuery Studio throught a Python request.
- In Cloud Storage, create a bucket
- In BigQuery Studio,
- create a Python notebook to transfer the Wikidata’s JSON feed to your bucket as JSON file
- create a dataset and table from the JSON file, with automatic detection of its scheme
- export the new table to Google Cloud Storage as a PARQUET file
- Download your files in Google Cloud Storage, and vizualise them locally
My conclusion
| Criteria | MS Azure | AWS Glue | GCP |
|---|---|---|---|
| Direct connection to the feed | No: from JSON feed to JSON file | Yes: work directly from the JSON feed | No: from JSON feed to JSON file |
| Data vizualisation | Yes: from input feed to output dataset | Yes: only output dataset | No: have to download output dataset |
| Easy to manipulate | Yes, but details to validate and deploy again and again | Yes, but still with some specific terms to learn | Yes, but a bunch of old tools in the way |
AWS Glue’s visual interface is much easier to understand than the multiple validations and publications needed in MS Azure Data Factory, or the estonishing non-web native manipulation in GCP.